Research
Our research focuses on how genetic information is reshuffled from one generation to the next through meiotic recombination. Meiotic recombination events cluster in 1-2 kb wide recombination hotspots. These hotspots are activated in trans by PR-domain containing 9 (PRDM9). PRDM9 has a DNA binding domain consisting of a variable number of C2H2 zinc-fingers, which defines DNA binding specificity to hotspot motifs. The zinc-finger domain shows accelerated evolution across diverse metazoan taxa and thus recombination landscapes overlap only poorly, even in closely related species, such as human and chimpanzee.
Prdm9 shows high intra-species variation in humans and mice. Crosses of mice where PRDM9 variants of parents are incompatible with each others DNA binding motifs will result in F1 male hybrid sterility, making Prdm9 a speciation gene. PRDM9 also has a SET domain with H3K4 tri-methylation activity, responsible for targeting the hotspot sites for recombination initiation. In Prdm9 knockout mice, recombination is instead initiated at existing H3K4 tri-methylation marks, for example at promoters, resulting in meiotic arrest. Lack of functional Prdm9 thus typically results in azoospermia and consequently infertility.
Current Research Projects
The mechanism of recombination in the absence of Prdm9

The mammalian family of Canidae, such as dogs and wolves, as well as several orders of birds and fish lack a functional copy of PRDM9. They are nevertheless able to complete meiosis successfully. The genomes of dogs as well as zebra finch and long-tailed finch even possess typical signatures of punctate historical recombination, present as hotspots of breakdown of linkage disequilibrium.
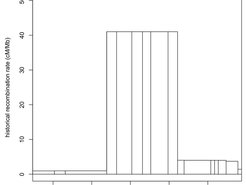
We are experts in investigating sites of historical recombination for de-novo recombination events using sperm-typing approaches. We are determining not only the frequency of recombination but also the placement of recombination breakpoints at the fine-scale, deciphering exactly where recombination events are distributed. In addition we use immune-fluorescent staining of meiotic proteins in cell spreads to assess meiotic timing as well as meiotic defects such as arrest and asynapsis (see banner image).
Comparing fine-scale recombination patterns of several animals with PRDM9 independent hotspot regulation, to organisms with PRDM9 regulated hotspots will give detailed insights into the difference between the dynamics and regulating factors of recombination in the absence of PRDM9, contributing to a better understanding of the function and role of PRDM9 in normal meiosis.
To achieve this aim, we are investigating numerous samples of Prdm9-deficient organisms, including several species of the Canidae family and members of the order of Passeriformes. Of course, we also have samples of organisms with Prdm9 regulated hotspots, mainly samples of several species and subspecies of mice across the genus Mus, such as Mus musculus musculus, Mus musculus domesticus, Mus spretus, Mus spicilegus as well as Apodemus uraliensis as an outgroup.
Recombination Landscape Evolution
Recombination landscapes evolve rapidly. PRDM9-regulated hotspots, at least, are suppressed by mutations that disrupt the recombination initiation motif that PRDM9 binds to. In individuals heterozygous for the mutation, the suppressing allele always gets over transmitted through biased gene conversion, eventually silencing the recombination hotspot at the population level.
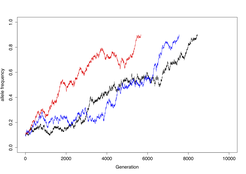
Additional biases, such as CG-biased gene conversion, do not affect crossovers or hotspot activity but nevertheless, generate meiotic drive and change genome composition over time. Recombination events are isolated from both orientations of recombination separately, using allele-specific PCR approaches. Comparing the frequency and distribution of reciprocal events uncovers incidences of bias and meiotic drive.
When observed, this information is used to predict the long-term evolutionary consequences via computer simulations.