Research Group Microbial Molecular Evolution
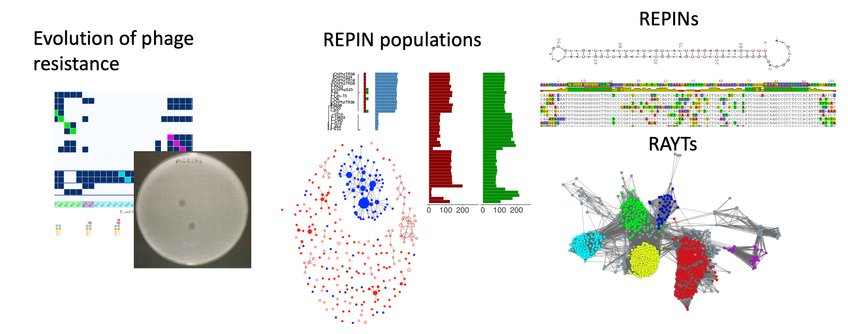
The evolution of phage resistance in bacteria
We are currently studying how E. coli C can become resistance to φX174 infection and in turn how φX174 can evolve to overcome E. coli C resistance. Currently we have characterized a large number of resistance mechanisms in E. coli C and how different resistance mechanisms can be overcome by different φX174 mutants. In the future we are interested in deepening our understanding of phage resistance and whether similar mechanisms could evolve in more complex environments such as the mammalian gut.
The ecology of REPIN populations
We have recently demonstrated that short repetitive sequences in bacterial genomes (REPINs) can be modeled as evolving populations (Genetics paper). Different to most other natural populations, an entire REPIN population evolves in a single bacterial genome. Hence by sequencing a bacterial genome we can attain a snap shot of the exact composition of a REPIN population. Similar to other natural populations, REPIN populations interact with their environment. We will study interactions such as competition and cooperation between populations, population decline and population growth. Our first study will focus on tracking the evolution of a single REPIN population through a number of P. fluorescens genomes and explore the alternative evolutionary paths this population has taken as the different P. fluorescens strains have diverged over time.
The functional significance of the REPIN-RAYT system for bacteria
As described above REPINs are short repetitive sequences in bacteria. RAYTs are the transposases that replicate REPINs (MGE paper and PLOS Genetics paper). Unlike selfish insertion sequences there is only a single RAYT that replicates one type of REPIN in a bacterial genome. RAYTs do not replicate themselves and are not horizontally transferred between bacteria. This means RAYTs have a function that is beneficial for the host bacterium, otherwise the gene would be easily lost over evolutionary time. Instead we find RAYTs in about 23% of all fully sequenced bacterial species (GBE paper).
We will perform a range of laboratory experiments to identify the functional significance of RAYTs.